Neil Bradman and Mark Thomas reveal the power of modern genetic analysis for exploring the role of fathers in human history.
In the beginning …
Genesis, chapter 5, records "the generations of Adam": Adam begat Seth, Seth begat Enosh, Enosh begat Kenan … down to Noah of the flood (Table 1). Translated into modern genetic terms, the account could read "Adam passed a copy of his Y chromosome (Figure 1) to Seth, Seth passed a copy of his Y chromosome to Enosh, Enosh passed a copy of his Y chromosome to Kenan" … and so on until Noah was born carrying a copy of Adam's Y chromosome. The Y chromosome is paternally inherited; human males have one while females have none. What is more, the Y chromosome a father passes to his son is, in large measure, an unchanged copy of his own.
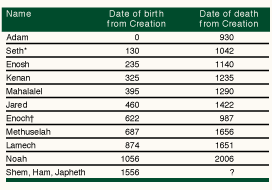
Table 1: before the flood: the generations of Adam according to the book of Genesis
But small changes (called polymorphisms) do occur, and this article describes how the correct interpretation of these changes in the Y chromosome, when passed down from generation to generation, can illuminate our understanding of human history. The study of Y chromosome differences is still in its infancy: until a few years ago, no polymorphisms had been discovered, and even today relatively few have been described. Studies recording the frequency of different combinations of polymorphisms (haplotypes) are even rarer.
The Y Chromosome
All human cells, other than mature red blood cells, possess a nucleus which contains the genetic material (DNA) arranged into 46 chromosomes, themselves grouped into 23 pairs.
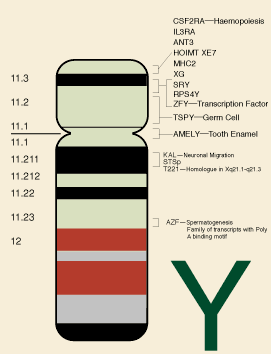
Figure 1: physical map of the Y chromosome indicating some known genes and gene regions
In 22 pairs, both members are essentially identical, one deriving from the individual's mother, the other from the father; such pairs are known as autosomes. The 23rd pair is different: while in females this pair has two like chromosomes called "X," in males it comprises one "X" and one "Y," two very dissimilar chromosomes. It is these chromosome differences which determine sex. During the production of sperm and eggs (gametes), the paired chromosomes separate so that each gamete ends up with only one member of each chromosome pair. However, before separation occurs, the paired autosomes swap pieces of their DNA with each other. In women, this exchange process also takes place between the two X chromosomes but, in men, unmatched X and Y chromosomes do not exchange DNA except at the ends of the two chromosomes; these are called the pseudoautosomal regions.
All eggs contain an X chromosome while sperm are of two types: those containing an X chromosome and those containing a Y. Fertilization restores the chromosomes to their normal paired condition. Thus, a Y sperm fertilizing the X egg (remember: all eggs carry only X because females have no Y chromosome) produces an XY zygote (cell produced by union of two gametes) which develops as male; fertilization by an X sperm gives rise to a female XX zygote. Since every male must possess a Y chromosome which can be inherited only from his father, a man's Y chromosome represents a unique record of his paternal inheritance.
There is another form of DNA which follows the female line of inheritance. Outside the nucleus, DNA contained in the energy-producing mitochondria is inherited only through the female line (the sperm's mitochondria are discarded in fertilization), providing its own unique record of female inheritance (Figure 2).
The chromosomes are mainly composed of DNA, that remarkable substance which constitutes "the book of life," the genetic instruction manual written in a four letter alphabet: A (adenine), T (thymine), C (cytosine) and G (guanine). It is these letters we must read to uncover the history recorded in the 60 million letters of the Y chromosome. In the copying of DNA from one generation to the next, mistakes, though rare, are sometimes made. Since the Y chromosome, unlike the autosomes in the other 22 pairs, does not exchange letters with a partner (except for the comparatively small pseudoautosomal regions), those mistakes are the only changes which are passed on to the next generation.
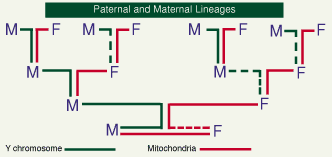
Figure 2: the different transmission paths of genetic material: Y chromosomes exclusively paternal, mitochondrial DNA entirely maternal
DNA can be classified into two categories: genic, in which the "letters" are involved in the production of the proteins responsible for most of the physical characteristics of each individual organism, and so-called junk which seems to have little apparent purpose. Most (perhaps 98 percent) of the Y chromosome is junk and changes in it are selected neither for nor against. So they are passed on - a record of an event which had no effect on the life of the man in whom the change occurred nor, indeed, on the life of his descendants.
Imagine an island in which over many generations the population numbers remain constant. Some men have sons, some do not: those with sons pass on their Y chromosomes while the Ys of the others are lost to history. It is easy to show that eventually the descendants of only one of the original Y chromosomes (in many copies) survive in the population because, once a particular line dies out, it never reappears. In the Y chromosome's passage through the generations, changes occur randomly in its junk DNA and so the Y chromosomes of the contemporary population retain a record of their passage through time: they can reveal the paternal genealogy of their owners and the relationships between different groups of individuals. For us as genetic anthropologists, that island is the whole world and we are the current generation whose genetic history is open for study.
Chromosome Changes
Changes that do occur from generation to generation are of four types:
- indels - insertions into or deletions of the DNA at particular locations on the chromosome. One insertion particularly useful in population studies is the YAP, which stands for "Y chromosome alu polymorphism." Alu is a sequence of approximately 300 letters (base pairs) which has inserted itself into a particular region of the DNA. There have been some half a million alu insertions in human DNA; YAP is one of the more recent.
- snips - are "single nucleotide polymorphisms" in which a particular nucleotide (an A, for example) is changed (perhaps into a G). Stable indels and snips are relatively rare and, in the case of the latter, so infrequent that it is reasonable to assume they have occurred at any particular position in the genome only once in the course of human evolution. Snips and stable alus have been termed "unique event polymorphisms" (UEPs).
Two other polymorphisms complete the marker set which can be used to unravel all Y chromosome history:
- microsatellites are short sequences of nucleotides (such as GATA) repeated over and over again a variable number of times in tandem. The specific number of repeats in a particular variant (or allele) usually remains unchanged from generation to generation but changes do sometimes occur and the number of repeats may increase or decrease.
It is usually assumed that increases or decreases in the number of repeats take place in single steps, for instance from nine repeats to ten, but whether decreases in number are as common as increases has not been established. Changes in microsatellite length occur much more frequently than new UEPs arise. What is more, while we can reasonably assume that a UEP has arisen only once, the number of repeat units in a microsatellite may have changed many times along a paternal lineage.
- the fourth polymorphism category is minisatellites, extensively studied by Mark Jobling at the University of Leicester. Unlike microsatellites, in which the repeated sequences are short (often no more than 3 or 4 nucleotides), in minisatellites they are normally 10-60 base pairs long and the number of repeats often extends to several dozen. Changes during the copying process take place more frequently in minisatellites than in microsatellites and the mechanisms may be different in the two cases.
In using polymorphisms to study changes over time, we are fortunate in having markers which change at different rates. Perhaps we can think of the UEPs as the hour hand, the microsatellite polymorphisms as the minute hand and the minisatellites as a sweep second hand of the evolutionary clock. Because most of the Y chromosome does not exchange DNA with a partner, a further benefit of using it to study evolution is that all the markers are joined one to another along its entire length. Such linkage of markers means that a haplotype constructed from a number of different markers records the evolutionary history of the particular Y chromosome on which they are all located.
To illustrate this, imagine (as, in fact, once did happen) that many years ago the Y chromosome of a particular man acquired the YAP insert; we call him YAP+. That man had one or more sons, sons who carried the YAP+ insert on their Y chromosomes. In turn, at least one of the man's sons (a grandson of the original YAP+) had one or more of his own sons … and so the process continued with the number of male offspring increasing until there were many men who had Y chromosomes with YAP+. Imagine further that in one such descendant (as, once again, did in fact happen), an A at one particular place on the Y chromosome was exchanged for a G and that the man in whom this took place also gave rise to an unbroken line of male descendants. There were now individuals in the population with the haplotype YAP+A and others with YAP+G, as well as those without the YAP insert whom we might designate as YAP-A. One of the Y chromosome microsatellites (DYS19) is found in various lengths (alleles) called 11, 12, 13, 14, etc. Thus, the haplotype of a man descended from the individual in whom A was mis-copied to G (who was, in turn, a descendant of the man in whom the YAP+ was inserted), and who has a DYS19 polymorphism of length 14, is described as YAP+ sY81(G) DYS19(14). Haplotypes can be used to construct trees describing the evolutionary history of the Y chromosome; in such evolutionary trees, UEPs provide the trunk and branches, and microsatellites the twigs.
The human Y chromosome has of course an ancestry predating the species it inhabits; haplotypes can also be used to draw evolutionary trees describing the relationships of the Y chromosomes of other primates.
Typing the Y
Unfortunately, identifying the haplotype of any particular individual is not as easy as reading the letters on a printed page. Analytical techniques are developing rapidly but a great deal of work is still involved. Obtaining the DNA itself is now relatively simple. Although it can be extracted from blood, cheek cells are just as good and can be collected simply by taking a mouth swab. The cell membranes are disrupted and DNA purified by one of a number of standard procedures. Small pieces of DNA called primers, copies of the nucleotide sequences located on both sides of a polymorphic site, are added to a mixture of the original DNA together with individual nucleotides (A, T, C and G) plus an enzyme called Taq, and the mixture heated and cooled 30-40 times so that multiple copies of the polymorphic loci can be produced (for details of this procedure, the Polymerase Chain Reaction [PCR], see Science Spectra No. 5, p. 50, 1996). The different polymorphic loci are distinguished from each other by their chain lengths, which can be measured using an automatic DNA sequencer (Figure 3).
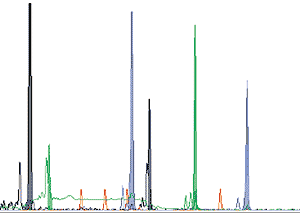
Figure 3: gene scan output of microsatellite DNA analysis from a single individual. The microsatellite peaks are sorted by size, the different colours representing different microsatellites. The small red peaks are size markers; the black peaks near the left-hand end are spurious DNA products resulting from the PCR process. Ordinate: quantity of DNA; abscissa: size of DNA fragment
Once typing is complete, haplotypes may be constructed and differences between them evaluated. Using reasonable assumptions about the rates at which different types of mutations occur, one can estimate a date for the most recent common ancestor (MRCA) of any two or more Y chromosomes, i.e. of any two living individuals. For example, Mike Hammer at the University of Arizona, having sequenced 2,400 bases in the same Y chromosome region from 16 ethnically diverse humans and four chimpanzees, was able to date the common ancestral human Y chromosome at 188,000 years with a 95 percent confidence interval from 51,000 to 411,000 years. The YAP+ insert was dated to 141,000 years before present, with a 95 percent confidence interval of 29,000 years to 340,000 years. In a different study published in the same issue of Nature, Whitfield et al in Cambridge interpreted data from sequencing 18,300 bases obtained from five ethnically diverse humans and a chimpanzee to give an MRCA time of between 37,000 and 49,000 years before present; in other words, those five men had a common great, great … great-grandfather some 40,000 or 50,000 years ago.
Where a significant number of individuals (usually more than 50) in a population have been analyzed, the frequency of occurrence of different haplotypes can be used both to distinguish populations and to shed light on the sub-structures within a population.
Not only does a new UEP arise on a particular Y chromosome at a particular time and in a particular person, it begins its history in the company of certain pre-existing microsatellite alleles. When a new UEP arises in a certain man, perhaps microsatellite DYS19 has a particular length. As the new UEP is copied from generation to generation, so too is DYS19. The UEP does not change but, albeit not very often, DYS19 does,perhaps increasing, perhaps decreasing in length. The longer the time since the UEP arose, the greater will be the number of different DYS19 alleles. Such a process differentiates one population from another; all things being equal, the more closely two populations display common haplotype frequencies, the more closely related is their biological history likely to be. Three examples illustrate these possibilities.
The South African Lemba
The Lemba are a black Southern African Bantu-speaking population who assert Jewish ancestry. Claiming Jewish origins is not an unusual phenomenon: the myth of the lost tribes is a powerful story and many groups have either claimed to be the descendants of one or other of the tribes or have been put forward for that honor.
The Lemba make no such grandiose claim but their oral history records an origin "in the north" as craftsmen in metalwork who traveled south to trade. Professor Mathivha of the University of the North, himself a Lemba, suggests an origin amongst Jewish traders in the Yemen. In a migration such as this, men and women may not have been equally represented amongst the travelers. In fact, Lemba tradition has it that at one time men left behind at a trading station received news of the fall of their homeland, took local wives, settled down and developed into the groups living today. If that, or something like it, were actually the case, we should expect to see more evidence of a non-Bantu origin in the paternally inherited Y chromosomes of the Lemba than in either their maternally-inherited mitochondrial DNA or their mixed autosomes. Intriguingly, half or more of the Lemba Y chromosomes are reported to have a Semitic origin although it is uncertain whether they were inherited from Jewish or Arab traders. Future interpretation will depend, in part, on the extent to which, if at all, it proves possible to differentiate the Y chromosomes of Jewish and Arab populations.
The Jewish Priests
A study in collaboration with Karl Skorecki, a nephrologist at the Rambam Medical Centre in Haifa, and Mike Hammer, looked at Jewish priests (Cohanim; singular Cohen). Cohanim are not the same as Rabbis; the latter are appointed functionaries while members of the priestly class inherit their position through the male line. In biblical tradition, Aaron (Figure 5), the brother of Moses, was the first priest: God awarded the priesthood to him - and his sons. In other words, the priesthood passes with the Y chromosome and there is no legitimate way in which a non-priest can become a priest. We reasoned that if oral tradition had been faithfully maintained, with the priesthood being passed, by and large, from father to son throughout the generations, an island of Y chromosomes of Jewish priests would have been created set within, but separated from, a sea of non-priests.
Applying what we knew about Y chromosome polymorphisms, two markers were selected for analysis. The first was a UEP which, we postulated, should, if the oral tradition had been maintained, be more homogeneous within the priestly group than in other Jews. This would be so because the Jewish population as a whole is likely to contain a variety of Y chromosomes contributed by men from many different communities converting to Judaism. Furthermore, since a Jew is defined as the son of a Jewess, the Y chromosomes, even of non-Jews, could enter the Jewish population. We also selected a microsatellite known to have a number of common alleles because, we argued, over time the microsatellite would have evolved independently in both the priestly Y chromosomes and those of the non-priests. In addition, as we have just noted, the non-priest Y chromosome pool would have been augmented by new entrants. As a consequence, we anticipated that the distribution of allele frequencies in the two populations might well be different. Of course, there are likely to have been some mistakes over time - men who forgot their father's priestly status, cases of false paternity and so on but, if the oral tradition had been maintained, a difference between the two populations should be observable.
Having constructed haplotypes using the YAP+, a UEP and the microsatellite DYS19, we noted the frequency of each haplotype in putative priests (Cohanim) and non-priests in both the Ashkenazic and non-Ashkenazic communities who, prior to the creation of the modern State of Israel, had occupied different geographical regions for 500 years or more. Using a simple statistical test (known as the ![]() 2 test), it was possible to distinguish the priestly from the non-priestly populations in both communities, suggesting that the oral tradition had indeed been maintained.
2 test), it was possible to distinguish the priestly from the non-priestly populations in both communities, suggesting that the oral tradition had indeed been maintained.
In Ancient Times
So far we have considered only the analysis of DNA obtained from our contemporaries and suggested ways in which we might deduce past history from an interpretation of those data. However, there are direct routes to the past: DNA can be extracted from ancient remains (see "Digging into the Past with DNA" in Science Spectra No. 6, p. 64, 1996) and human Y chromosomes from the Neolithic have now been typed. Marina Faerman and her colleagues at the Hadassah Medical School in Jerusalem adopted this approach to identify the sex of infant skeletons too young and incomplete to be classified in any other way. They took advantage of the fact that the amelogenin gene exists in two forms, the one on the X chromosome being different in length from the one on Y. Small portions of cautical bones, cranial bones and teeth were crushed to powder and decalcified; DNA was purified, copied by PCR using primers flanking the region, and the size of the products was measured by a technique known as agarose gel electrophoresis. Since Y chromosomes yield fragments 218 base pairs long while X chromosome products contain 330 base pairs, they should be clearly distinguishable: if the specimen yields the shorter gene, it must come from a Y chromosome fragment and thus from a male.
Would that the process were as straightforward as it sounds! In ancient remains, DNA is often degraded so that continuous fragments are no longer present and cannot be copied. In addition, substances may be present which inhibit both purification and amplification. But Faerman overcame these difficulties and obtained positive results from 18 separate specimens.
A further problem she faced was the risk of incorrectly identifying a skeleton as female when really it was male. This could arise if the Y chromosome DNA were so degraded that copies could not be made of the relevant 218-base pair fragment but were successfully obtained from the 330-base pair X chromosome sequence present in the same sample. Classification of a skeleton as male requires both products to be identified while a female skeleton should be expected to yield only the longer fragment. Faerman and her colleagues argued that the risk of misclassification was low since the longer X chromosome sequence would be more likely to be degraded than the shorter Y chromosome fragment. In addition, experiments undertaken under controlled conditions indicated that false positive readings are more likely than false negatives.
We Ourselves
On a personal note, we must say that there is something moving in feeding into a database one's own detailed Y chromosome haplotype and finding in that database a dozen or so relatives of the paternal line, members of far-flung populations, men whom one has never met, who know nothing of one's own existence but who, nevertheless, share a common ancestor not so long ago. The father of one of the author's daughter's best friends turned out to have a Y chromosome matching the author's own although neither knew in detail how they might be paternally related. The case of the other author, who probably derives from ancient Celtic stock, is even more remarkable. His Y chromosome haplotype has thus far been found only in two other individuals: one is his prospective Swedish father-in-law, the other a native of Turkey. But then, if we go back far enough, all men are not only born equal but are paternally related.
Glossary
ALLELE one of a number of alternative forms that can occupy a given genetic locus on a chromosome
AUTOSOME any chromosome other than the sex chromosomes
HAPLOTYPE the set of alleles borne on one of a pair of homologous chromosomes
POLYMORPHISM the existence within a species or population of different forms
Suggested Reading
- Cavalli-Sforza, L.L. and Cavalli-Sforza, F. The Great Human Diasporas: the History of Diversity and Evolution Addison-Wesley Publishing Co., Reading, MA (1995).
- Jobling, M.A. and Tyler-Smith, C. Fathers and Sons: the Y Chromosome and Human Evolution Trends in Genetics, 11, 449-456 (1995)
- Skorecki, K. et al Y Chromosomes of the Jewish Priests Nature, 385, 32 (1997)